Library : A library represents data generated from a single run of sequencing a sample. For example, in a RNA-seq experiment with 3 repeats each of control and treatment will constitute 6 libraries irrespective of the fact, if they are single end or paired end sequenced. If you have sequenced same sample multiple times and want us to treat it as a single sample by combining the different runs, we will consider it as a single library, except in case genome assemblies where data from each runs will be treated as different libraries.
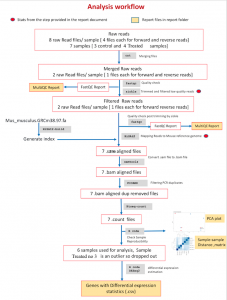
RNAseq analysis scope: The analysis listed typically include – the quality control, alignment, read analysis post alignment (package:qualimap) relevant statistical models for DE analysis, and database pulls for associating their function. We will perform reproducibility of biological replicates using PCA and sample-sample distance matrix. The final report will include expression of detected genes with their fold change values, relevant figures and code used in the study. A sample analysis workflow is shown below
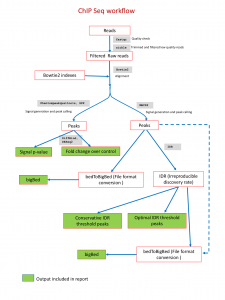
ChIP-Seq Analysis scope: Quality evaluation of raw reads (fastqc/multiqc), quality control (Sickle/Trimmomatic), alignment of short reads to reference (HISAT2), Peak identification (MACS2), Feature extraction: Consensus sequence, Genomic features near peaks. Differential peak analysis. ENCODE recommended workflow guidelines will be followed.