The Cluster
- What is a Cluster?
- Our Cluster
- Connecting to the Cluster
- Xming/Xquartz
- Putty Configuration
- Connecting from outside the UConn domain
- Windows
- Linux
- BBC cluster layout
- Cluster Etiquette
- Monitoring User Quota
Interacting with SGE
- Interactive Sessions
- qlogin
- qrsh
- Batch Job
- Composing Script
- Queue on Cluster and their Status
- All Queue
- Highpri Queue
- Lowpri Queue
- Highmem Queue
Submitted Jobs
The Cluster
What is a Cluster? A desktop or a laptop, in most cases, is inadequate for analysis of large scale datasets (e.g. Genomics, transcriptomics) or to run simulations (e.g. protein dockings, weather forecast). They lack both the processing power and the RAM to carry out these analyses. This limitation can be overcome by joining many of these machines (computers) in a predefined architecture/configuration so that they act as a single unit with enhanced computational power and enabled to share resources. This is the basic concept of a high performance cluster. A cluster consists of a set connected computers that work together so that they can be viewed as a single system. Each computer unit in a cluster is referred as ‘node’.
The components of a cluster are usually connected through fast local area networks (“LAN”), with each node running its own instance of an operating system. Clusters improve performance and availability of a single computer. The benefits of clusters include low cost, elasticity and being able to run jobs anytime, anywhere.
Our Cluster The BBC Bioinformatics Facility Dell cluster consists of numerous nodes, each of which is a separate machine with its own hard drive and 2 processors, each having 4 cores. Normally, a single job requires one core, so a single node could run 8 separate analyses simultaneously. A job is always started on the head node, which is the node you are using when you log in to the cluster. Your job may or may not run on the head node; it is up to the scheduler software to decide where your job will run. The software that manages your run is known as the Sun Grid Engine, or SGE for short. It schedules jobs to run in a cluster environment and utilizes system resources in the most efficient way possible. SGE consists of several programs (qsub, qstat, etc.) that you will use to get your run started and check in on its progress.
In order to display graphics from the cluster, we need a software that allows one to use Linux graphical applications remotely. Xming and Xquartz are the display options available for windows and mac respectively. (Download and install it on your local computers)
Windows: Xming
Mac: Xquatz
NOTE: Start the X Server on your machine (Xming/Xquartz), each time you reboot your PC or whenever you want to use X Windows. Once enabled, Xming will appear in your system tray as a black X with an orange circle around the middle.
To log-in to the head node of the cluster run the command Mac or Linux terminal:
ssh your_username@bbcsrv3.biotech.uconn.edu or ssh –X your_username@bbcsrv3.biotech.uconn.edu
-X is to enable X11 forwarding to enable graphics display from cluster.
Windows users will need to use an SSH client to connect to the cluster. Please refer to the PuTTY guide on tutorial section.
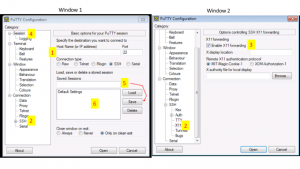
Install Putty and configure for use.
Putty Configuration steps.
Open Putty it will open window1.
- Provide host name e.g. your_username@bbcsrv3.biotech.uconn.edu
- Expand SSH tab and select X11 (shown in window2)
- Enable X11 forwarding by selecting it. (window2)
- Scroll up the left panel and select Session.(window1)
- Name your session e.g. BBC_cluster and click save tab to save.
- Your session name should appear in saved sessions.
Double click on your session name to connect to server with SSH session.
SSH logins from outside the UConn domain SSH logins from outside the UConn domain (UConn-Storrs, UConn-Avery Point, UCHC) must now use the UConn VPN client prior to connecting through your usual SSH client.
Windows and OS X: Junos Pulse:
Please see http://remoteaccess.uconn.edu/vpn-overview/connect-via-vpn-client/ for instructions on setting up a Junos Pulse VPN client.
Linux: Openconnect (ex. using Ubuntu):
Step 1: Install required libraries and tools sudo apt-get install vpnc sudo chmod a+x+r -R /etc/vpnc sudo apt-get install libxml2 libxml2-dev gettext make libssl-dev pkg-config libtool autoconf git Step 2: Download, build and install openconnect git clone git://git.infradead.org/users/dwmw2/openconnect.git cd openconnect autoreconf -iv ./configure make sudo make install Step 3: Connect to the VPN over terminal openconnect --juniper sslvpn.uconn.edu
Now openconnect should prompt you to enter your netID and password. Upon successful login, openconnect should confirm an ssl connection has been established. Once this happens, create a new terminal window and enter the ssh command you use to connect to the server.
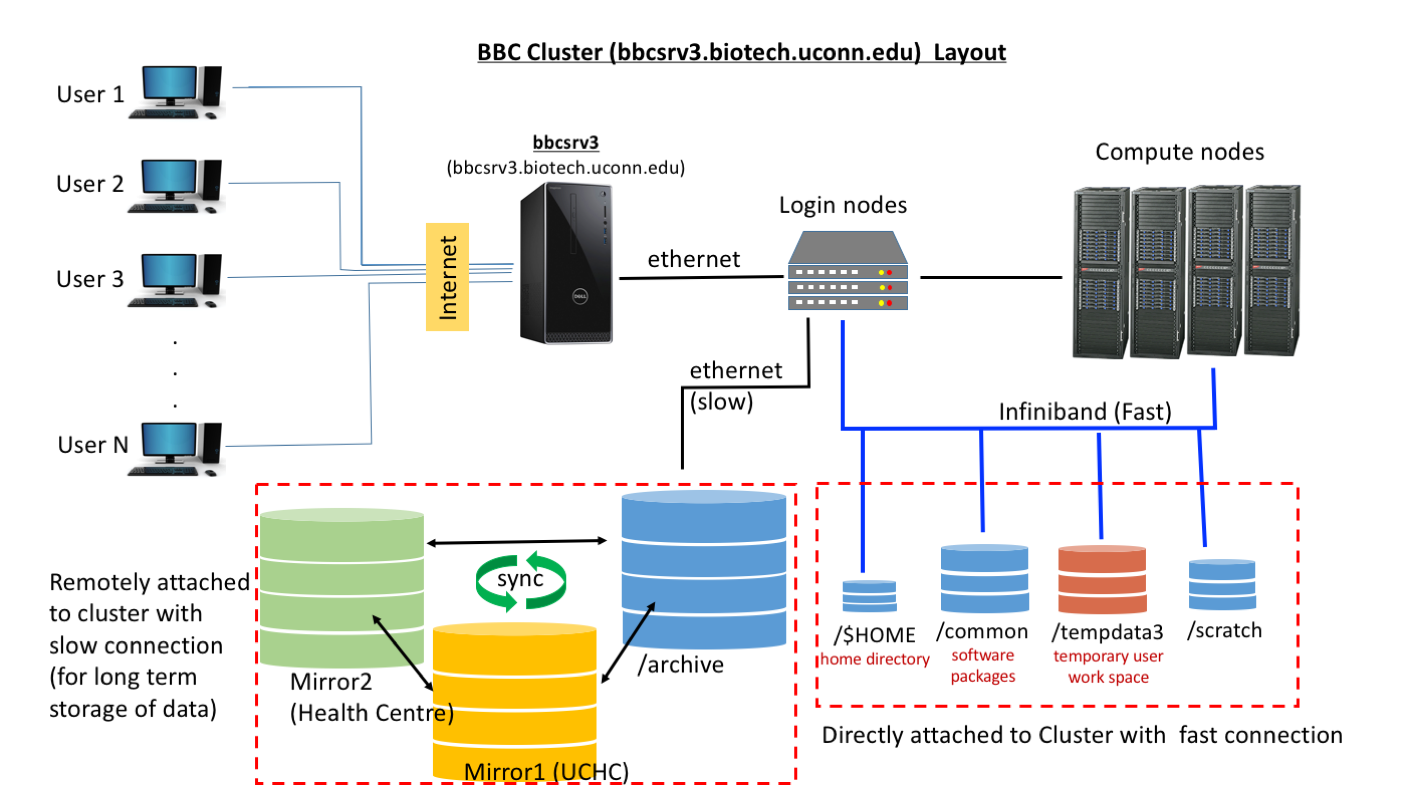
From user perspective there are 5 important components and each one with specific properties.
- Head Node (bbcsrv3): Head node is the gateway for the users to communicate with the cluster. The principal role of Head node is to provide management and job scheduling services to the cluster.
- NOTE: HEAD NODE SHOULD NOT BE USED FOR COMPUTATIONAL PURPOSES. IF CLUSTER BECOMES SLOW OR NON RESPONSIVE DUE TO JOBS RUNNING ON HEAD NODE, SUCH JOBS WILL BE TERMINATED WITHOUT WARNING.
- Login Nodes and compute nodes: User can login on these nodes using ‘qlogin’ command and can have interactive sessions. They are ideal for interactive sessions for testing scripts. To use them type “qlogin’ at command prompt and “exit” when you want to end your session.
- Fast connection directories: Your home directory, /common , /tempdata3 and /tempdata2 are mounted/connected on the cluster in a configuration such that they are optimized for fast data access by compute nodes and head node./$HOME : Your home directory is relatively small space and may be suffice only to hold small amount of data. Use it wisely.
- /common : This directory holds databases (http://bioinformatics.uconn.edu/databases/ ) and bioinformatics softwares. Avoid using it at all cost. It’s a limited space and is required for storing common resources.
- /tempdata3: This directory is designed for users to store their files while they are performing analysis on them.
- NOTE: THIS IS MENT FOR TEMPORARLY HOLDING YOUR FILES WHILE THEY ARE USED IN ANALYSIS. THIS DRIVE IS NOT BACKED UP AND A CRASH MAY LEAD TO LOSS OF DATA BEYOND RECOVERY. IT IS WISE TO MOVE YOUR FILES AS SOON AS THEIR USE IS OVER.
- /tempdata2: This disk space smaller than the /tempdata3, but it can also can be used by users
- /archive : This drive has big storage capacity and is backed up at two other centers. This is ideal for long term storage of data files and archiving. However due to its mount and network connection to cluster, accessing data is not very fast.SOLUTION: Copy your files to /tempdadta3 for analysis and after you are done copy results back to archive and clear space (on /tempdata3) for other users.
- Never run anything on the head node of a cluster.
- Keep track of what you are running and refrain from using all nodes at the same time.
- Run and write your output files on scratch drive or /tempdata3. Then copy the file back to your home directory or /archive.
To check how much space is used/available on your home directory:
quota -u username
Interacting with SGE
Once a you logged on BBC cluster, by default you are directed to your home directory. At this point you are still on head node and any command or computation you perform is executed on head node, something that we do not recommended. For computing purpose user has two options
- Running an interactive session (qlogin or qrsh).
- Submitting a script (qsub).
An interactive job starts when you log onto the system and ends when you log off. Even the activities on head node is an interactive session. In interest of all users, it is recommended that for all interactive sessions users should logon to nodes, with help of following commands
qlogin qrsh
Once you type qlogin you get logged on to one of the nodes (e.g.: in it is compute-1-0 node)
$ qlogin
Your job 236491 ("QLOGIN") has been submitted
waiting for interactive job to be scheduled ...
Your interactive job 236491 has been successfully scheduled.
Establishing /opt/gridengine/bin/rocks-qlogin.sh session to host compute-1-0.local ...
Last login: Wed Feb 1 17:37:27 2017 from bbcsrv3.local
Rocks Compute Node
Rocks 6.1 (Emerald Boa)
Profile built 09:01 01-Nov-2016
Kickstarted 09:17 01-Nov-2016
[user@compute-1-0 ~]$
qrsh will connect to node (e.g. in this case it is compute-1-16 node)
$ qrsh [user@compute-1-16 ~]$
‘exit’ command will move you back on head node.
If resources are available, the job gets started immediately. Otherwise, the user is informed about the lack of resources and the job gets aborted. Jobs are always passed onto the available executing hosts. Records of each job’s progress through the system are kept and reported when requested (qstat). qrsh is similar to qlogin by submitting an interactive job to the queuing system. However, qrsh establishes a rsh connection with the remote host. qlogin uses the current terminal for user I/O.
SSH directly into a specific node on the cluster
First, view the current load on the cluster with the qhost command. Then determine which nodes satisfy your computational needs (number of CPU, MEMTOT). Of those nodes, select the node that has the lowest current LOAD and MEMUSE.
After a node has been selected connect to the node with the terminal command:
ssh HOSTNAME # where HOSTNAME is the hostname of the node
For example, if the screenshot below represented the current load on the server, and a highpri queue was required, then compute-2-2 or compute-2-3 would be selected since they are both empty.
Batch Job A batch job requires little or no interaction between the user and the system. It consists of a predefined group of processing actions. If resources are available, job gets started immediately. Otherwise, the user is informed about the lack of resources and job gets abandoned.
A sample template of script and example scripts are at the end of this document to help you compose your own.
Some key points to consider:
Depending on the computational resource required by your software, one of the queues on the cluster has to be specified. The decision relies on the fact whether the task is CPU or memory intensive.
Software’s that creates large tmp files and hold them in RAM will often be memory intensive e.g. (k-mer estimation, transcriptome assemblies, genome assemblies etc). Other tasks are generally CPU intensive and specifying more cores will speed up the process (e.g, Bowtie, STAR, RSEM, SICKLE, TOPHAT etc).
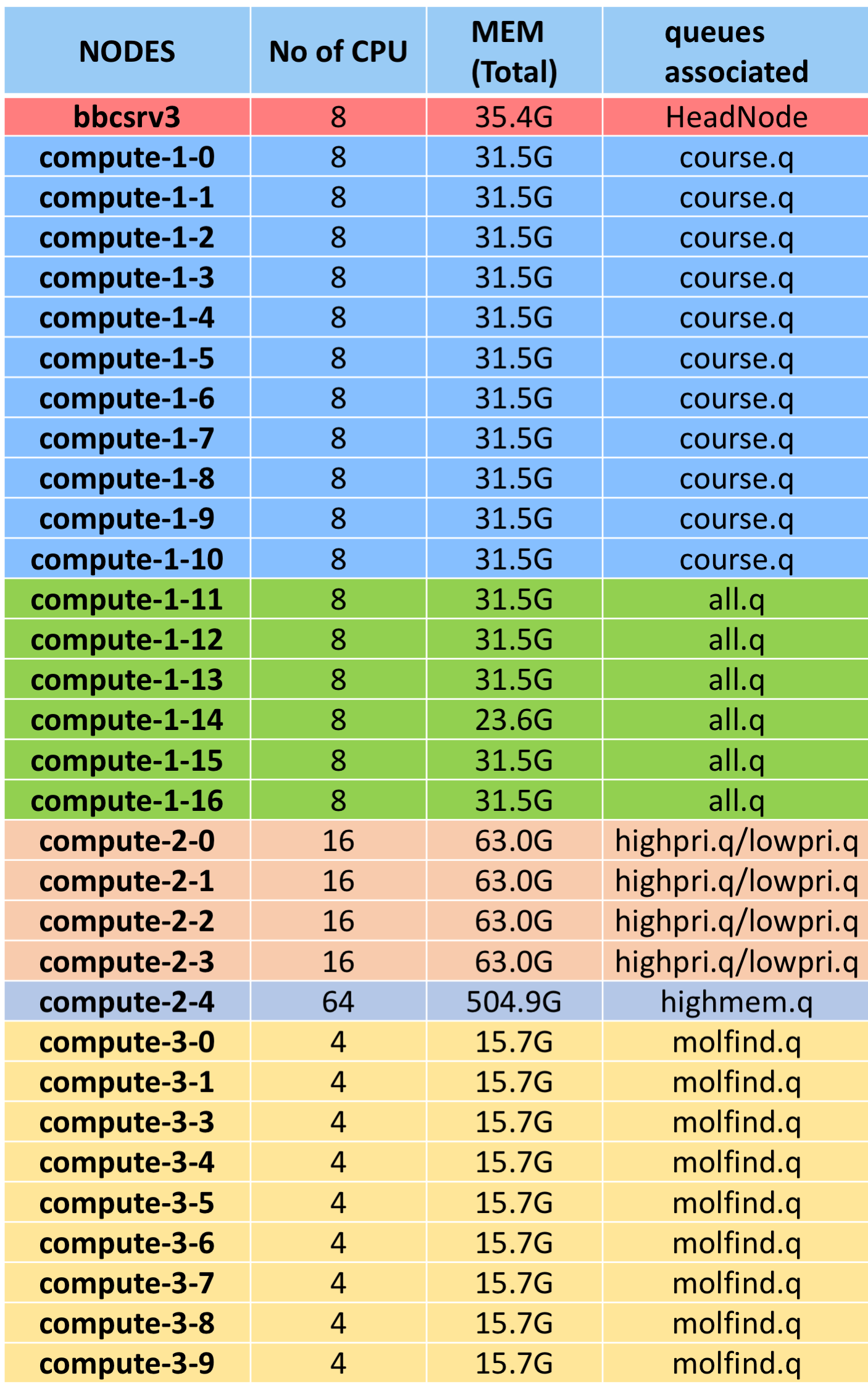
Queue on cluster and their status:
$ qstat –g c
CLUSTER QUEUE CQLOAD USED RES AVAIL TOTAL aoACDS cdsuE -------------------------------------------------------------------------------- all.q 0.41 78 0 54 132 0 0 highmem.q 0.44 64 0 0 64 0 0 highpri.q 0.65 64 0 0 64 0 0 lowpri.q 0.65 0 0 0 64 64 0 molfind.q 0.00 0 0 36 36 0 0
Column 1: Queue Name Column 3: No. processors in use. Column 5: Processors available to use. Column 6: Total processors designated to the queue.
All queue: This is the default queue for a job submitted using qsub and when queue is not specified.
Highpri queue: This queue is not available to the general user as the nodes were purchased using funds from outside the BBC. The queue can be used to run priority jobs. Specify the use of this queue with the following option in an SGE script:
#$ -q highpri.q
The total memory available is 63 gigabytes on each compute node.
Lowpri queue: This queue is available for general use, runs on the same four nodes as the highpri.q, but can only run single-threaded jobs. In addition, jobs running in the lowpri.q can be preempted by jobs in the highpri.q. Specify the use of this queue with the following option in an SGE script:
#$ -q lowpri.q
The total memory available is 63 gigabytes on each compute node.
Highmem queue: This queue should be used if a job requires more memory and CPU than offered by the regular queue. Specify the use of this queue with the following option in an SGE script:
#$ -q highmem.q
Please note that this queue limits each user to a maximum of 16 threads. The total memory available is 505 gigabytes.
DETAILS OF NODES AND QUEUE ASSOCIATED
Submitted Jobs
NOTE: Please make sure that the software you are running supports multi-threading. Usually this information is available in manual. If it does not, then it will not matter how many cores you specify, the software will still be running on one core and other cores will be idle which could have been used by other user.
Once you have composed your script please submit it using qsub.
QSub qsub is used for both single and multiple node jobs:
qsub scriptname.sh
Job Status A submitted job will;
- Wait in the queue
- Will be executed
- Be completed and leave the SGE scheduling system
qstat
This can be used if the job is in either (1) or (2) state. The command will inform the user if the job is still waiting or if it has started being executed. For state (1): SGE treats every node as a queue. If resources are unavailable, the job will not be executed. Once the resources do become available, the job will leave the queue and move on to state 2. qstat will provide information about the state of the job. qdel can be used to delete jobs from the queue. For state (2):
Once the job is submitted, check its status using qstat.
qstat
# this will list the jobs submitted by the user and their status. Below is the example where qstat lists the 2 jobs submitted by user vsingh.
job-ID prior name user state submit/start at queue slots ja-task-ID ----------------------------------------------------------------------------------------------------------------- 236976 0.50500 dwnld_1 vsingh r 02/14/2017 09:43:20 all.q@compute-1-3.local 1 237005 0.60500 rep_us vsingh r 02/14/2017 11:50:00 highpri.q@compute-2-3.local 16
Column 1: JobID a unqiue ID to identify the job Column 3: Name of the Job Column 4: Job owner’s username Column 5: Status of the job. r : Running R : Restarted qw : Waiting in queue t : Transferring dr : Job ending s,S: Suspended eqw: Error queuing job Column 6: Job submission time Column 7: Queue in which job is running Column 8: No of cores used by the job
Tip:
Obtaining error from a qsub job in ‘eqw’ status. This will help you troubleshoot the problem.
qstat –j jobID | grep error
This is used to monitor the job’s status (e.g. time and memory consumption)
qstat –j job_number
This will give time and memory consumed information For state (3):
qstat -j job_number | grep mem
This is the only command that may be able to tell you about the past jobs by referring to a database of past usage.
qstat -j job_number
The following are variations on the qstat command:
qstat -f #full listing for all users qstat -u username #specific to user qstat -f -u username #detailed information
Job Removal This command will remove the specified jobs that are waiting to be run, or kill jobs that are already running.
qdel job number
List of jobs:
qdel job number1 job number2 job number3
All jobs running or queueing under a given username:
qdel -u username
qhost
This command is used to view the load on SGE. The output includes the architecture (ARCH), the number of CPUs (NCPU), the current load (LOAD), the total memory (MEMTOT), the memory currently in use (MEMUSE) and the swap space (SWAPTO) for each node. To show the queues available for each node, add the parameter -q. To show the jobs currently running on each node, add the parameter -j.
HOSTNAME ARCH NCPU LOAD MEMTOT MEMUSE SWAPTO SWAPUS ------------------------------------------------------------------------------- global - - - - - - - bbcsrv3 linux-x64 8 28.84 35.4G 28.0G 2.0G 2.0G compute-1-0 linux-x64 8 3.39 31.5G 672.9M 2.0G 1.6M compute-1-1 linux-x64 8 10.01 31.5G 1.5G 2.0G 410.8M compute-1-10 linux-x64 8 3.55 31.5G 14.6G 2.0G 1.9G compute-1-11 linux-x64 8 2.07 31.5G 15.6G 2.0G 14.4M compute-1-12 linux-x64 8 6.06 31.5G 1.8G 2.0G 16.2M compute-1-13 linux-x64 8 4.82 31.5G 2.8G 2.0G 143.9M compute-1-14 linux-x64 8 1.09 23.6G 718.8M 2.0G 17.5M compute-1-15 linux-x64 8 2.55 31.5G 853.1M 2.0G 314.4M compute-1-16 linux-x64 8 10.07 31.5G 1.4G 2.0G 16.9M compute-1-2 linux-x64 8 12.10 31.5G 1.8G 2.0G 16.7M compute-1-3 linux-x64 8 4.77 31.5G 2.2G 2.0G 17.5M compute-1-4 linux-x64 8 3.00 31.5G 7.3G 2.0G 2.0G compute-1-5 linux-x64 8 4.57 31.5G 3.4G 2.0G 20.2M compute-1-6 linux-x64 8 8.14 31.5G 1.9G 2.0G 16.4M compute-1-7 linux-x64 8 1.02 31.5G 641.4M 2.0G 14.5M compute-1-8 linux-x64 8 8.80 31.5G 1.9G 2.0G 559.3M compute-1-9 linux-x64 8 1.12 31.5G 488.3M 2.0G 16.1M compute-2-0 linux-x64 16 1.00 63.0G 998.2M 2.0G 13.0M compute-2-1 linux-x64 16 1.00 63.0G 997.5M 2.0G 15.5M compute-2-2 linux-x64 16 0.00 63.0G 846.3M 2.0G 14.4M compute-2-3 linux-x64 16 7.19 63.0G 7.0G 2.0G 14.2M compute-2-4 linux-x64 64 29.49 504.9G 46.9G 2.0G 33.3M compute-3-0 linux-x64 4 0.02 15.7G 350.8M 2.0G 0.0 compute-3-1 linux-x64 4 0.00 15.7G 351.4M 2.0G 0.0 compute-3-3 linux-x64 4 0.00 15.7G 350.6M 2.0G 0.0 compute-3-4 linux-x64 4 0.00 15.7G 353.6M 2.0G 0.0 compute-3-5 linux-x64 4 0.00 15.7G 352.3M 2.0G 0.0 compute-3-6 linux-x64 4 0.00 15.7G 353.4M 2.0G 0.0 compute-3-7 linux-x64 4 0.00 15.7G 351.9M 2.0G 0.0 compute-3-8 linux-x64 4 0.00 15.7G 352.2M 2.0G 0.0 compute-3-9 linux-x64 4 0.01 15.7G 350.6M 2.0G 0.0
SSH directly into a specific node on the cluster
First, view the current load on the cluster with the qhost command. Then determine which nodes satisfy your computational needs (number of CPU, MEMTOT). Of those nodes, select the node that has the lowest current LOAD and MEMUSE.
After a node has been selected connect to the node with the terminal command:
ssh HOSTNAME # where HOSTNAME is the hostname of the node
For example, if the screenshot below represented the current load on the server, and a highpri queue was required, then compute-2-2 or compute-2-3 would be selected since they are both empty.
Often a loss of connection or closing of the terminal terminates the process you were working on, e.g. moving or coping large files. Screen command is very useful to avoid such situations. Once the terminal is activated using screen command anything in that terminal stays running even if you log out.
screen # It will activate the terminal screen –S test # It will activate the screen and name it as test ctrl + a + d # It detaches the screen and hide it. screen –r # List of all detached screens. screen –r test # This will reattach the screen name test screen –wipe test # wipes a screen when it is dead screen -S test -p 0 -X quit # Terminates the screen with name test